Jupyter Snippet NP ch14-code-listing
Jupyter Snippet NP ch14-code-listing
Chapter 14: Statistical modelling
Robert Johansson
Source code listings for Numerical Python - Scientific Computing and Data Science Applications with Numpy, SciPy and Matplotlib (ISBN 978-1-484242-45-2).
import statsmodels.api as sm
import statsmodels.formula.api as smf
import statsmodels.graphics.api as smg
import patsy
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy import stats
import seaborn as sns
Statistical models and patsy formula
np.random.seed(123456789)
y = np.array([1, 2, 3, 4, 5])
x1 = np.array([6, 7, 8, 9, 10])
x2 = np.array([11, 12, 13, 14, 15])
X = np.vstack([np.ones(5), x1, x2, x1*x2]).T
X
array([[ 1., 6., 11., 66.],
[ 1., 7., 12., 84.],
[ 1., 8., 13., 104.],
[ 1., 9., 14., 126.],
[ 1., 10., 15., 150.]])
beta, res, rank, sval = np.linalg.lstsq(X, y)
/Users/rob/miniconda3/envs/py3.6/lib/python3.6/site-packages/ipykernel_launcher.py:1: FutureWarning: `rcond` parameter will change to the default of machine precision times ``max(M, N)`` where M and N are the input matrix dimensions.
To use the future default and silence this warning we advise to pass `rcond=None`, to keep using the old, explicitly pass `rcond=-1`.
"""Entry point for launching an IPython kernel.
beta
array([-5.55555556e-01, 1.88888889e+00, -8.88888889e-01, -8.88178420e-16])
data = {"y": y, "x1": x1, "x2": x2}
y, X = patsy.dmatrices("y ~ 1 + x1 + x2 + x1*x2", data)
y
DesignMatrix with shape (5, 1)
y
1
2
3
4
5
Terms:
'y' (column 0)
X
DesignMatrix with shape (5, 4)
Intercept x1 x2 x1:x2
1 6 11 66
1 7 12 84
1 8 13 104
1 9 14 126
1 10 15 150
Terms:
'Intercept' (column 0)
'x1' (column 1)
'x2' (column 2)
'x1:x2' (column 3)
type(X)
patsy.design_info.DesignMatrix
np.array(X)
array([[ 1., 6., 11., 66.],
[ 1., 7., 12., 84.],
[ 1., 8., 13., 104.],
[ 1., 9., 14., 126.],
[ 1., 10., 15., 150.]])
df_data = pd.DataFrame(data)
y, X = patsy.dmatrices("y ~ 1 + x1 + x2 + x1:x2", df_data, return_type="dataframe")
X
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
model = sm.OLS(y, X)
result = model.fit()
result.params
Intercept -5.555556e-01
x1 1.888889e+00
x2 -8.888889e-01
x1:x2 -6.661338e-16
dtype: float64
model = smf.ols("y ~ 1 + x1 + x2 + x1:x2", df_data)
result = model.fit()
result.params
Intercept -5.555556e-01
x1 1.888889e+00
x2 -8.888889e-01
x1:x2 -6.661338e-16
dtype: float64
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 1.000
Model: OLS Adj. R-squared: 1.000
Method: Least Squares F-statistic: 2.807e+27
Date: Mon, 06 May 2019 Prob (F-statistic): 3.56e-28
Time: 15:42:33 Log-Likelihood: 149.18
No. Observations: 5 AIC: -292.4
Df Residuals: 2 BIC: -293.5
Df Model: 2
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept -0.5556 9.62e-14 -5.77e+12 0.000 -0.556 -0.556
x1 1.8889 3.59e-13 5.26e+12 0.000 1.889 1.889
x2 -0.8889 1.22e-13 -7.27e+12 0.000 -0.889 -0.889
x1:x2 -6.661e-16 1.13e-14 -0.059 0.958 -4.92e-14 4.79e-14
==============================================================================
Omnibus: nan Durbin-Watson: 0.023
Prob(Omnibus): nan Jarque-Bera (JB): 0.391
Skew: 0.250 Prob(JB): 0.822
Kurtosis: 1.724 Cond. No. 6.86e+17
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
[2] The smallest eigenvalue is 1.31e-31. This might indicate that there are
strong multicollinearity problems or that the design matrix is singular.
/Users/rob/miniconda3/envs/py3.6/lib/python3.6/site-packages/statsmodels/stats/stattools.py:72: ValueWarning: omni_normtest is not valid with less than 8 observations; 5 samples were given.
"samples were given." % int(n), ValueWarning)
beta
array([-5.55555556e-01, 1.88888889e+00, -8.88888889e-01, -8.88178420e-16])
from collections import defaultdict
data = defaultdict(lambda: np.array([1,2,3]))
patsy.dmatrices("y ~ a", data=data)[1].design_info.term_names
['Intercept', 'a']
patsy.dmatrices("y ~ 1 + a + b", data=data)[1].design_info.term_names
['Intercept', 'a', 'b']
patsy.dmatrices("y ~ -1 + a + b", data=data)[1].design_info.term_names
['a', 'b']
patsy.dmatrices("y ~ a * b", data=data)[1].design_info.term_names
['Intercept', 'a', 'b', 'a:b']
patsy.dmatrices("y ~ a * b * c", data=data)[1].design_info.term_names
['Intercept', 'a', 'b', 'a:b', 'c', 'a:c', 'b:c', 'a:b:c']
patsy.dmatrices("y ~ a * b * c - a:b:c", data=data)[1].design_info.term_names
['Intercept', 'a', 'b', 'a:b', 'c', 'a:c', 'b:c']
data = {k: np.array([]) for k in ["y", "a", "b", "c"]}
patsy.dmatrices("y ~ a + b", data=data)[1].design_info.term_names
['Intercept', 'a', 'b']
patsy.dmatrices("y ~ I(a + b)", data=data)[1].design_info.term_names
['Intercept', 'I(a + b)']
patsy.dmatrices("y ~ a*a", data=data)[1].design_info.term_names
['Intercept', 'a']
patsy.dmatrices("y ~ I(a**2)", data=data)[1].design_info.term_names
['Intercept', 'I(a ** 2)']
patsy.dmatrices("y ~ np.log(a) + b", data=data)[1].design_info.term_names
['Intercept', 'np.log(a)', 'b']
z = lambda x1, x2: x1+x2
patsy.dmatrices("y ~ z(a, b)", data=data)[1].design_info.term_names
['Intercept', 'z(a, b)']
Categorical variables
data = {"y": [1, 2, 3], "a": [1, 2, 3]}
patsy.dmatrices("y ~ - 1 + a", data=data, return_type="dataframe")[1]
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
patsy.dmatrices("y ~ - 1 + C(a)", data=data, return_type="dataframe")[1]
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
data = {"y": [1, 2, 3], "a": ["type A", "type B", "type C"]}
patsy.dmatrices("y ~ - 1 + a", data=data, return_type="dataframe")[1]
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
patsy.dmatrices("y ~ - 1 + C(a, Poly)", data=data, return_type="dataframe")[1]
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
Linear regression
np.random.seed(123456789)
N = 100
x1 = np.random.randn(N)
x2 = np.random.randn(N)
data = pd.DataFrame({"x1": x1, "x2": x2})
def y_true(x1, x2):
return 1 + 2 * x1 + 3 * x2 + 4 * x1 * x2
data["y_true"] = y_true(x1, x2)
e = np.random.randn(N)
data["y"] = data["y_true"] + e
data.head()
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
fig, axes = plt.subplots(1, 2, figsize=(8, 4))
axes[0].scatter(data["x1"], data["y"])
axes[1].scatter(data["x2"], data["y"])
fig.tight_layout()
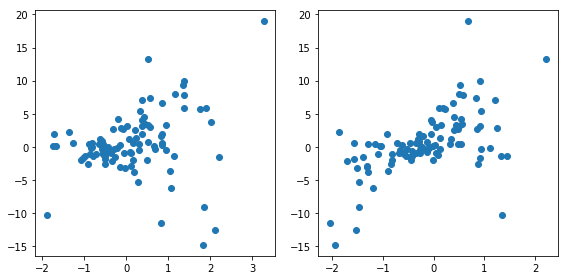
data.shape
(100, 4)
model = smf.ols("y ~ x1 + x2", data)
result = model.fit()
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.380
Model: OLS Adj. R-squared: 0.367
Method: Least Squares F-statistic: 29.76
Date: Mon, 06 May 2019 Prob (F-statistic): 8.36e-11
Time: 15:42:50 Log-Likelihood: -271.52
No. Observations: 100 AIC: 549.0
Df Residuals: 97 BIC: 556.9
Df Model: 2
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 0.9868 0.382 2.581 0.011 0.228 1.746
x1 1.0810 0.391 2.766 0.007 0.305 1.857
x2 3.0793 0.432 7.134 0.000 2.223 3.936
==============================================================================
Omnibus: 19.951 Durbin-Watson: 1.682
Prob(Omnibus): 0.000 Jarque-Bera (JB): 49.964
Skew: -0.660 Prob(JB): 1.41e-11
Kurtosis: 6.201 Cond. No. 1.32
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
result.rsquared
0.3802538325513254
result.resid.head()
0 -3.370455
1 -11.153477
2 -11.721319
3 -0.948410
4 0.306215
dtype: float64
z, p = stats.normaltest(result.resid.values)
p
4.6524990253009316e-05
result.params
Intercept 0.986826
x1 1.081044
x2 3.079284
dtype: float64
fig, ax = plt.subplots(figsize=(8, 4))
smg.qqplot(result.resid, ax=ax)
fig.tight_layout()
fig.savefig("ch14-qqplot-model-1.pdf")
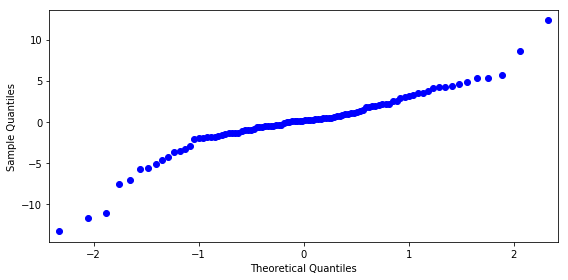
model = smf.ols("y ~ x1 + x2 + x1*x2", data)
result = model.fit()
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.955
Model: OLS Adj. R-squared: 0.954
Method: Least Squares F-statistic: 684.5
Date: Mon, 06 May 2019 Prob (F-statistic): 1.21e-64
Time: 15:42:53 Log-Likelihood: -140.01
No. Observations: 100 AIC: 288.0
Df Residuals: 96 BIC: 298.4
Df Model: 3
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 0.8706 0.103 8.433 0.000 0.666 1.076
x1 1.9693 0.108 18.160 0.000 1.754 2.185
x2 2.9670 0.117 25.466 0.000 2.736 3.198
x1:x2 3.9440 0.112 35.159 0.000 3.721 4.167
==============================================================================
Omnibus: 2.950 Durbin-Watson: 2.072
Prob(Omnibus): 0.229 Jarque-Bera (JB): 2.734
Skew: 0.327 Prob(JB): 0.255
Kurtosis: 2.521 Cond. No. 1.38
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
result.params
Intercept 0.870620
x1 1.969345
x2 2.967004
x1:x2 3.943993
dtype: float64
result.rsquared
0.9553393745884368
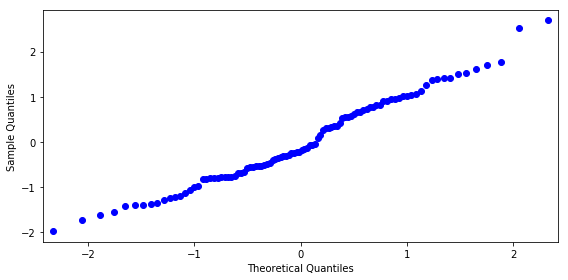
z, p = stats.normaltest(result.resid.values)
p
0.22874710482505126
fig, ax = plt.subplots(figsize=(8, 4))
smg.qqplot(result.resid, ax=ax)
fig.tight_layout()
fig.savefig("ch14-qqplot-model-2.pdf")
x = np.linspace(-1, 1, 50)
X1, X2 = np.meshgrid(x, x)
new_data = pd.DataFrame({"x1": X1.ravel(), "x2": X2.ravel()})
y_pred = result.predict(new_data)
y_pred.shape
(2500,)
y_pred = y_pred.values.reshape(50, 50)
fig, axes = plt.subplots(1, 2, figsize=(12, 5), sharey=True)
def plot_y_contour(ax, Y, title):
c = ax.contourf(X1, X2, Y, 15, cmap=plt.cm.RdBu)
ax.set_xlabel(r"$x_1$", fontsize=20)
ax.set_ylabel(r"$x_2$", fontsize=20)
ax.set_title(title)
cb = fig.colorbar(c, ax=ax)
cb.set_label(r"$y$", fontsize=20)
plot_y_contour(axes[0], y_true(X1, X2), "true relation")
plot_y_contour(axes[1], y_pred, "fitted model")
fig.tight_layout()
fig.savefig("ch14-comparison-model-true.pdf")
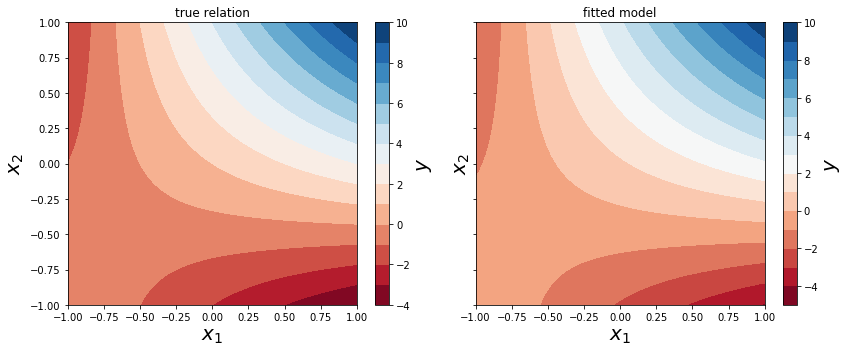
Datasets from R
dataset = sm.datasets.get_rdataset("Icecream", "Ecdat")
dataset.title
'Ice Cream Consumption'
print(dataset.__doc__)
+----------+-----------------+
| Icecream | R Documentation |
+----------+-----------------+
Ice Cream Consumption
---------------------
Description
~~~~~~~~~~~
four–weekly observations from 1951–03–18 to 1953–07–11
*number of observations* : 30
*observation* : country
*country* : United States
Usage
~~~~~
::
data(Icecream)
Format
~~~~~~
A time serie containing :
cons
consumption of ice cream per head (in pints);
income
average family income per week (in US Dollars);
price
price of ice cream (per pint);
temp
average temperature (in Fahrenheit);
Source
~~~~~~
Hildreth, C. and J. Lu (1960) *Demand relations with autocorrelated
disturbances*, Technical Bulletin No 2765, Michigan State University.
References
~~~~~~~~~~
Verbeek, Marno (2004) *A Guide to Modern Econometrics*, John Wiley and
Sons, chapter 4.
See Also
~~~~~~~~
``Index.Source``, ``Index.Economics``, ``Index.Econometrics``,
``Index.Observations``,
``Index.Time.Series``
dataset.data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 30 entries, 0 to 29
Data columns (total 4 columns):
cons 30 non-null float64
income 30 non-null int64
price 30 non-null float64
temp 30 non-null int64
dtypes: float64(2), int64(2)
memory usage: 1.0 KB
model = smf.ols("cons ~ -1 + price + temp", data=dataset.data)
result = model.fit()
print(result.summary())
OLS Regression Results
==============================================================================
Dep. Variable: cons R-squared: 0.986
Model: OLS Adj. R-squared: 0.985
Method: Least Squares F-statistic: 1001.
Date: Mon, 06 May 2019 Prob (F-statistic): 9.03e-27
Time: 15:43:04 Log-Likelihood: 51.903
No. Observations: 30 AIC: -99.81
Df Residuals: 28 BIC: -97.00
Df Model: 2
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
price 0.7254 0.093 7.805 0.000 0.535 0.916
temp 0.0032 0.000 6.549 0.000 0.002 0.004
==============================================================================
Omnibus: 5.350 Durbin-Watson: 0.637
Prob(Omnibus): 0.069 Jarque-Bera (JB): 3.675
Skew: 0.776 Prob(JB): 0.159
Kurtosis: 3.729 Cond. No. 593.
==============================================================================
Warnings:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
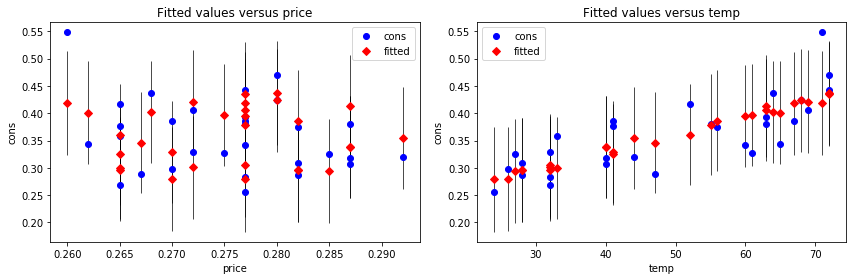
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
smg.plot_fit(result, 0, ax=ax1)
smg.plot_fit(result, 1, ax=ax2)
fig.tight_layout()
fig.savefig("ch14-regressionplots.pdf")
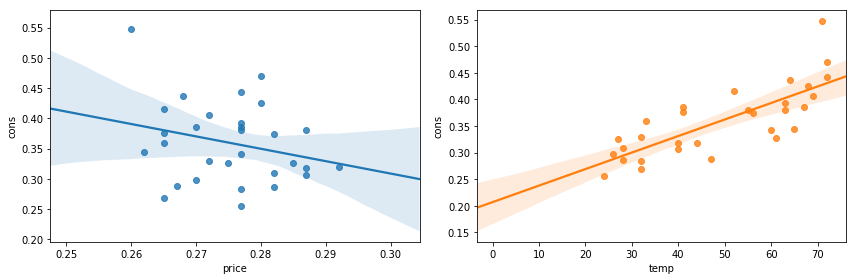
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
sns.regplot("price", "cons", dataset.data, ax=ax1);
sns.regplot("temp", "cons", dataset.data, ax=ax2);
fig.tight_layout()
fig.savefig("ch14-regressionplots-seaborn.pdf")
Discrete regression, logistic regression
df = sm.datasets.get_rdataset("iris").data
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 5 columns):
Sepal.Length 150 non-null float64
Sepal.Width 150 non-null float64
Petal.Length 150 non-null float64
Petal.Width 150 non-null float64
Species 150 non-null object
dtypes: float64(4), object(1)
memory usage: 5.9+ KB
df.Species.unique()
array(['setosa', 'versicolor', 'virginica'], dtype=object)
df_subset = df[(df.Species == "versicolor") | (df.Species == "virginica" )].copy()
df_subset = df[df.Species.isin(["versicolor", "virginica"])].copy()
df_subset.Species = df_subset.Species.map({"versicolor": 1, "virginica": 0})
df_subset.rename(columns={"Sepal.Length": "Sepal_Length", "Sepal.Width": "Sepal_Width",
"Petal.Length": "Petal_Length", "Petal.Width": "Petal_Width"}, inplace=True)
df_subset.head(3)
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
model = smf.logit("Species ~ Sepal_Length + Sepal_Width + Petal_Length + Petal_Width", data=df_subset)
result = model.fit()
Optimization terminated successfully.
Current function value: 0.059493
Iterations 12
print(result.summary())
Logit Regression Results
==============================================================================
Dep. Variable: Species No. Observations: 100
Model: Logit Df Residuals: 95
Method: MLE Df Model: 4
Date: Mon, 06 May 2019 Pseudo R-squ.: 0.9142
Time: 15:43:11 Log-Likelihood: -5.9493
converged: True LL-Null: -69.315
LLR p-value: 1.947e-26
================================================================================
coef std err z P>|z| [0.025 0.975]
--------------------------------------------------------------------------------
Intercept 42.6378 25.708 1.659 0.097 -7.748 93.024
Sepal_Length 2.4652 2.394 1.030 0.303 -2.228 7.158
Sepal_Width 6.6809 4.480 1.491 0.136 -2.099 15.461
Petal_Length -9.4294 4.737 -1.990 0.047 -18.714 -0.145
Petal_Width -18.2861 9.743 -1.877 0.061 -37.381 0.809
================================================================================
Possibly complete quasi-separation: A fraction 0.60 of observations can be
perfectly predicted. This might indicate that there is complete
quasi-separation. In this case some parameters will not be identified.
print(result.get_margeff().summary())
Logit Marginal Effects
=====================================
Dep. Variable: Species
Method: dydx
At: overall
================================================================================
dy/dx std err z P>|z| [0.025 0.975]
--------------------------------------------------------------------------------
Sepal_Length 0.0445 0.038 1.163 0.245 -0.031 0.120
Sepal_Width 0.1207 0.064 1.891 0.059 -0.004 0.246
Petal_Length -0.1703 0.057 -2.965 0.003 -0.283 -0.058
Petal_Width -0.3303 0.110 -2.998 0.003 -0.546 -0.114
================================================================================
Note: Sepal_Length and Sepal_Width do not seem to contribute much to predictiveness of the model.
model = smf.logit("Species ~ Petal_Length + Petal_Width", data=df_subset)
result = model.fit()
Optimization terminated successfully.
Current function value: 0.102818
Iterations 10
print(result.summary())
Logit Regression Results
==============================================================================
Dep. Variable: Species No. Observations: 100
Model: Logit Df Residuals: 97
Method: MLE Df Model: 2
Date: Mon, 06 May 2019 Pseudo R-squ.: 0.8517
Time: 15:43:13 Log-Likelihood: -10.282
converged: True LL-Null: -69.315
LLR p-value: 2.303e-26
================================================================================
coef std err z P>|z| [0.025 0.975]
--------------------------------------------------------------------------------
Intercept 45.2723 13.612 3.326 0.001 18.594 71.951
Petal_Length -5.7545 2.306 -2.496 0.013 -10.274 -1.235
Petal_Width -10.4467 3.756 -2.782 0.005 -17.808 -3.086
================================================================================
Possibly complete quasi-separation: A fraction 0.34 of observations can be
perfectly predicted. This might indicate that there is complete
quasi-separation. In this case some parameters will not be identified.
print(result.get_margeff().summary())
Logit Marginal Effects
=====================================
Dep. Variable: Species
Method: dydx
At: overall
================================================================================
dy/dx std err z P>|z| [0.025 0.975]
--------------------------------------------------------------------------------
Petal_Length -0.1736 0.052 -3.347 0.001 -0.275 -0.072
Petal_Width -0.3151 0.068 -4.608 0.000 -0.449 -0.181
================================================================================
params = result.params
beta0 = -params['Intercept']/params['Petal_Width']
beta1 = -params['Petal_Length']/params['Petal_Width']
df_new = pd.DataFrame({"Petal_Length": np.random.randn(20)*0.5 + 5,
"Petal_Width": np.random.randn(20)*0.5 + 1.7})
df_new["P-Species"] = result.predict(df_new)
df_new["P-Species"].head(3)
0 0.995472
1 0.799899
2 0.000033
Name: P-Species, dtype: float64
df_new["Species"] = (df_new["P-Species"] > 0.5).astype(int)
df_new.head()
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
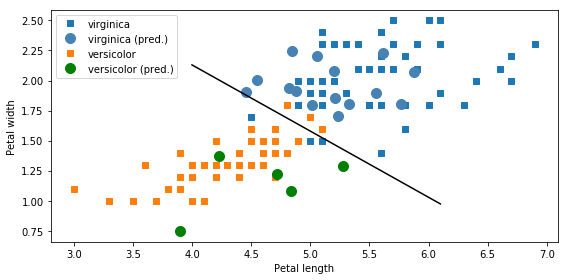
fig, ax = plt.subplots(1, 1, figsize=(8, 4))
ax.plot(df_subset[df_subset.Species == 0].Petal_Length.values,
df_subset[df_subset.Species == 0].Petal_Width.values, 's', label='virginica')
ax.plot(df_new[df_new.Species == 0].Petal_Length.values,
df_new[df_new.Species == 0].Petal_Width.values,
'o', markersize=10, color="steelblue", label='virginica (pred.)')
ax.plot(df_subset[df_subset.Species == 1].Petal_Length.values,
df_subset[df_subset.Species == 1].Petal_Width.values, 's', label='versicolor')
ax.plot(df_new[df_new.Species == 1].Petal_Length.values,
df_new[df_new.Species == 1].Petal_Width.values,
'o', markersize=10, color="green", label='versicolor (pred.)')
_x = np.array([4.0, 6.1])
ax.plot(_x, beta0 + beta1 * _x, 'k')
ax.set_xlabel('Petal length')
ax.set_ylabel('Petal width')
ax.legend(loc=2)
fig.tight_layout()
fig.savefig("ch14-logit.pdf")
Poisson distribution
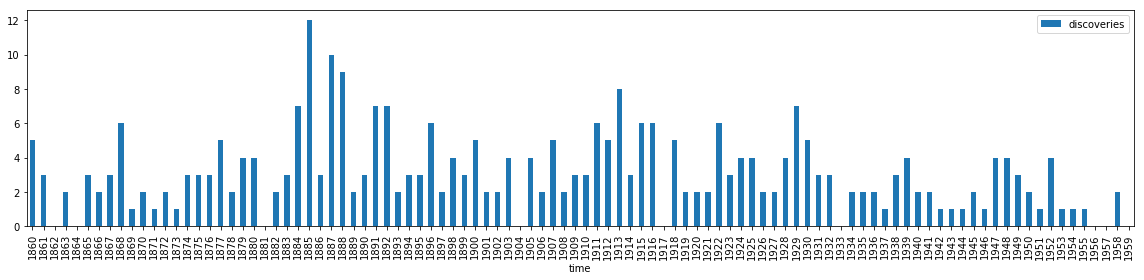
dataset = sm.datasets.get_rdataset("discoveries")
df = dataset.data.set_index("time").rename(columns={"value": "discoveries"})
df.head(10).T
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
fig, ax = plt.subplots(1, 1, figsize=(16, 4))
df.plot(kind='bar', ax=ax)
fig.tight_layout()
fig.savefig("ch14-discoveries.pdf")
model = smf.poisson("discoveries ~ 1", data=df)
result = model.fit()
Optimization terminated successfully.
Current function value: 2.168457
Iterations 1
print(result.summary())
Poisson Regression Results
==============================================================================
Dep. Variable: discoveries No. Observations: 100
Model: Poisson Df Residuals: 99
Method: MLE Df Model: 0
Date: Mon, 06 May 2019 Pseudo R-squ.: 0.000
Time: 15:43:21 Log-Likelihood: -216.85
converged: True LL-Null: -216.85
LLR p-value: nan
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 1.1314 0.057 19.920 0.000 1.020 1.243
==============================================================================
lmbda = np.exp(result.params)
X = stats.poisson(lmbda)
result.conf_int()
.dataframe tbody tr th {
vertical-align: top;
}
.dataframe thead th {
text-align: right;
}
X_ci_l = stats.poisson(np.exp(result.conf_int().values)[0, 0])
X_ci_u = stats.poisson(np.exp(result.conf_int().values)[0, 1])
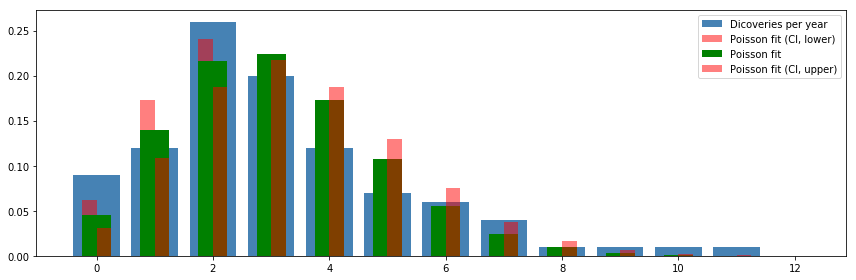
v, k = np.histogram(df.values, bins=12, range=(0, 12), normed=True)
/Users/rob/miniconda3/envs/py3.6/lib/python3.6/site-packages/ipykernel_launcher.py:1: VisibleDeprecationWarning: Passing `normed=True` on non-uniform bins has always been broken, and computes neither the probability density function nor the probability mass function. The result is only correct if the bins are uniform, when density=True will produce the same result anyway. The argument will be removed in a future version of numpy.
"""Entry point for launching an IPython kernel.
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.bar(k[:-1], v, color="steelblue", align='center', label='Dicoveries per year')
ax.bar(k-0.125, X_ci_l.pmf(k), color="red", alpha=0.5, align='center', width=0.25, label='Poisson fit (CI, lower)')
ax.bar(k, X.pmf(k), color="green", align='center', width=0.5, label='Poisson fit')
ax.bar(k+0.125, X_ci_u.pmf(k), color="red", alpha=0.5, align='center', width=0.25, label='Poisson fit (CI, upper)')
ax.legend()
fig.tight_layout()
fig.savefig("ch14-discoveries-per-year.pdf")
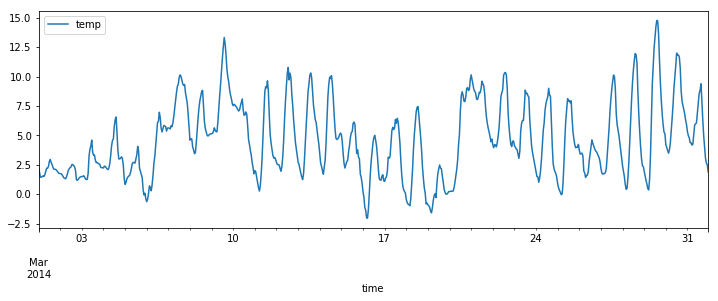
Time series
df = pd.read_csv("temperature_outdoor_2014.tsv", header=None, delimiter="\t", names=["time", "temp"])
df.time = pd.to_datetime(df.time, unit="s")
df = df.set_index("time").resample("H").mean()
df_march = df[df.index.month == 3]
df_april = df[df.index.month == 4]
df_march.plot(figsize=(12, 4));
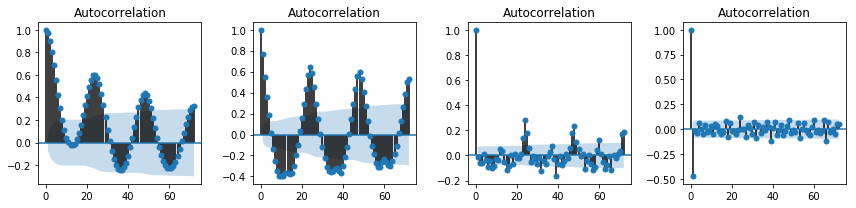
fig, axes = plt.subplots(1, 4, figsize=(12, 3))
smg.tsa.plot_acf(df_march.temp, lags=72, ax=axes[0])
smg.tsa.plot_acf(df_march.temp.diff().dropna(), lags=72, ax=axes[1])
smg.tsa.plot_acf(df_march.temp.diff().diff().dropna(), lags=72, ax=axes[2])
smg.tsa.plot_acf(df_march.temp.diff().diff().diff().dropna(), lags=72, ax=axes[3])
fig.tight_layout()
fig.savefig("ch14-timeseries-autocorrelation.pdf")
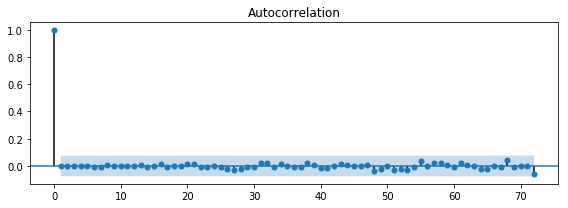
model = sm.tsa.AR(df_march.temp)
result = model.fit(72)
sm.stats.durbin_watson(result.resid)
1.9985623006352906
fig, ax = plt.subplots(1, 1, figsize=(8, 3))
smg.tsa.plot_acf(result.resid, lags=72, ax=ax)
fig.tight_layout()
fig.savefig("ch14-timeseries-resid-acf.pdf")
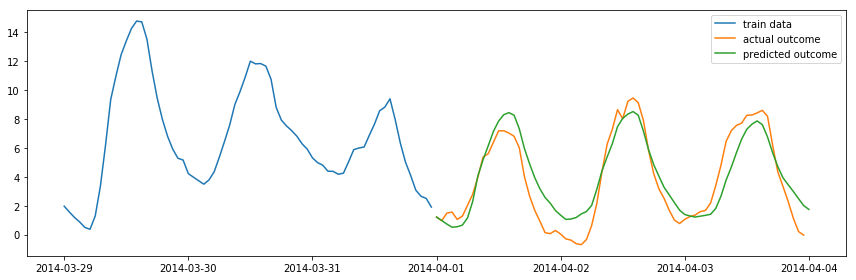
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.plot(df_march.index.values[-72:], df_march.temp.values[-72:], label="train data")
ax.plot(df_april.index.values[:72], df_april.temp.values[:72], label="actual outcome")
ax.plot(pd.date_range("2014-04-01", "2014-04-4", freq="H").values,
result.predict("2014-04-01", "2014-04-4"), label="predicted outcome")
ax.legend()
fig.tight_layout()
fig.savefig("ch14-timeseries-prediction.pdf")
/Users/rob/miniconda3/envs/py3.6/lib/python3.6/site-packages/statsmodels/tsa/base/tsa_model.py:336: FutureWarning: Creating a DatetimeIndex by passing range endpoints is deprecated. Use `pandas.date_range` instead.
freq=base_index.freq)
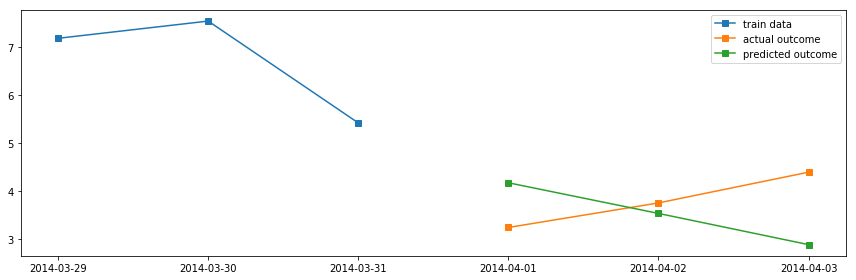
# Using ARMA model on daily average temperatures
df_march = df_march.resample("D").mean()
df_april = df_april.resample("D").mean()
model = sm.tsa.ARMA(df_march, (4, 1))
result = model.fit()
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
ax.plot(df_march.index.values[-3:], df_march.temp.values[-3:], 's-', label="train data")
ax.plot(df_april.index.values[:3], df_april.temp.values[:3], 's-', label="actual outcome")
ax.plot(pd.date_range("2014-04-01", "2014-04-3").values,
result.predict("2014-04-01", "2014-04-3"), 's-', label="predicted outcome")
ax.legend()
fig.tight_layout()
Versions
%reload_ext version_information
%version_information numpy, matplotlib, pandas, scipy, statsmodels, patsy